Dominant subtype switch in avian influenza viruses during 2016-2019 in China - PubMed
. 2020 Nov 20;11(1):5909.
doi: 10.1038/s41467-020-19671-3.
Yuhai Bi 1 2 3 , Shanqin Li 5 , Guanghua Fu 6 , Tao Jin 7 , Cheng Zhang 8 9 , Yongchun Yang 10 , Zhenghai Ma 9 , Wenxia Tian 11 , Jida Li 12 , Shuqi Xiao 13 , Liqiang Li 7 , Renfu Yin 14 , Yi Zhang 12 , Lixin Wang 15 , Yantao Qin 16 , Zhongzi Yao 17 , Fanyu Meng 4 , Dongfang Hu 18 , Delong Li 19 , Gary Wong 20 21 , Fei Liu 8 , Na Lv 8 , Liang Wang 8 , Lifeng Fu 8 , Yang Yang 5 , Yun Peng 5 , Jinmin Ma 7 , Kirill Sharshov 22 , Alexander Shestopalov 22 , Marina Gulyaeva 22 , George F Gao 8 5 23 24 , Jianjun Chen 17 , Yi Shi 8 5 , William J Liu 24 , Dong Chu 25 , Yu Huang 6 , Yingxia Liu 23 , Lei Liu 23 , Wenjun Liu 8 5 , Quanjiao Chen 26 , Weifeng Shi 27 28
Affiliations
- PMID: 33219213
- PMCID: PMC7679419
- DOI: 10.1038/s41467-020-19671-3
Free PMC article
Dominant subtype switch in avian influenza viruses during 2016-2019 in China
Yuhai Bi et al. Nat Commun. .
Free PMC article
Abstract
We have surveyed avian influenza virus (AIV) genomes from live poultry markets within China since 2014. Here we present a total of 16,091 samples that were collected from May 2016 to February 2019 in 23 provinces and municipalities in China. We identify 2048 AIV-positive samples and perform next generation sequencing. AIV-positive rates (12.73%) from samples had decreased substantially since 2016, compared to that during 2014-2016 (26.90%). Additionally, H9N2 has replaced H5N6 and H7N9 as the dominant AIV subtype in both chickens and ducks. Notably, novel reassortants and variants continually emerged and disseminated in avian populations, including H7N3, H9N9, H9N6 and H5N6 variants. Importantly, almost all of the H9 AIVs and many H7N9 and H6N2 strains prefer human-type receptors, posing an increased risk for human infections. In summary, our nation-wide surveillance highlights substantial changes in the circulation of AIVs since 2016, which greatly impacts the prevention and control of AIVs in China and worldwide.
Conflict of interest statement
The authors declare no competing interests.
Figures

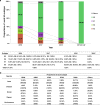
a Map of the AIV sampling sites and isolation rates in LPMs. AIV surveillance sites in 37 cities (indicated by black dots) of 23 provinces or municipalities or minority autonomous regions in China are divided into seven different regions: North (Inner Mongolia, NM and Jilin, JL; orange), East-Central (Shanxi, SX; Ningxia, NX; Shandong, SD; Shaanxi, SaX and Henan, HeN; light green), South-Central (Anhui, AH; Hunan, HuN; Jiangxi, JX and Fujian, FJ; yellow), Yangtze River Delta (Jiangsu, JS and Zhejiang, ZJ; pink), South-West (Sichuan, SC; Chongqing, CQ; Yunnan, YN; and Guizhou, GZ; orange red), South (Guangxi, GX; Guangdong, GD and Hainan, HaN; dark green), and West (Xinjiang, XJ; Qinghai, QH; and Xizang, XZ; light purple). The red portion in each pie chart indicates the isolation rate of AIV in this region. The standard map was downloaded from Ministry of Natural Resources of the People’s Republic of China (
http://bzdt.ch.mnr.gov.cn/), and the collection sites of LPMs in our study were marked on the map using ArcGIS. b AIV positive rates of the present study (2016–19) and the previous study in 2014–16. The regions included North, East-Central, South-Central, Yangtze River Delta, South-West, and South. The numbers on the column represent the AIV isolation rate. c Subtype proportions of AIVs in the pure isolates with a single HxNy subtype. d The proportion of HA and NA from the impure isolates containing over two HA or NA subtypes. Source data are provided as a Source Data file.
a The proportion of various HA subtypes of pure isolates with a single HxNy subtype isolated between 2016 and 2019. The major prevalent subtypes include H5, H6, H7, and H9. H5 viruses include H5N6 and H5N8; H6 viruses include H6N2, H6N6, and H6N8; H7 viruses include H7N2, H7N3, H7N6, H7N7, and H7N9; H9 viruses include H9N2, H9N6, and H9N9. b Host species distributions of H5N6, H6N6, H7N3, H7N9, H9N2, and other subtypes. Source data are provided as a Source Data file.

a Phylogenetic tree of the HA gene of H9Ny AIVs. b Phylogenetic tree of the NA gene of H9N2 viruses. Viruses are marked with different colors according to the collection dates (before 2017: blue violet, in 2017: orange, in 2018: light green, and in 2019: light blue). Both trees are rooted using CK/BJ/1/1994(H9N2). The light blue and red triangles represent H9N6 and H9N9 viruses, respectively. All blue dots in the phylogenetic trees (a) represent H9N2 and H9N9 strains used for the receptor-binding test in this study. The labels with gray lines indicate all of the H9 isolates sequenced in this study.

a Phylogenetic tree of the HA gene of H5 AIVs. The orange, light blue, red, and light green lines in the tree represent viruses described in this study isolated from 2016, 2017, 2018, and 2019, respectively. The dark blue lines represent the reference strains previously reported by Bi et al.. The subtrees marked with a pink and light blue background represent the major lineage (Clade 2.3.4.4d) and the minor lineage (Clade 2.3.4.4a), respectively. The purple lines represent other reference strains from the Influenza Virus Resource at NCBI and the GISAID databases. b Phylogenetic tree of the HA gene of H7 AIVs. The subtrees marked with a pink and light blue background represent H7 strains belonging to the Yangtze River Delta lineage and Pearl River Delta lineage, respectively. The subtree of the H7N9 HPAIVs previously analyzed by Quan et al. is marked with the blue background on the upper right. The orange, light blue, and red lines of the tree represent strains isolated from 2016, 2017, and 2018, respectively. The subtree displayed in the dashed frame on the upper right included the HA genes of 33 H7N3 isolates in this study. The dotted lines represent H7N2 (n = 3), H7N6 (n = 3), and H7N8 (n = 1) viruses identified in this study. c The topology of the HA tree of H7 AIVs was shown at the bottom-right, with dots represent 160 H7 AIV strains identified in our surveillance during 2016–19. All blue dots in the phylogenetic trees (a, b) represent the H5 and H7 strains used for receptor-binding test in this study. In addition, the red pentagrams represent the H5/H7 bivalent vaccine strains, A/chicken/Guizhou/4/2013(H5N1) and A/pigeon/Shanghai/S1069/2013(H7N9), respectively.
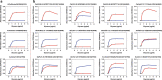
a A/Anhui/1/2013(H7N9) was used as a reference for comparison with the tested H7 strains. Two human strains, A/California/04/2009(H1N1) and A/Vietnam/1194/2004(H5N1), were used as controls. b Receptor-binding properties of the representative AIV strains to human (α2-6-SA) and avian (α2-3-SA) receptors were tested using the solid-phase direct binding assay with trisaccharide receptors. Red and blue lines represent human- and avian-type receptors, respectively. Two replications presented similar results and the mean values were shown. Source data are provided as a Source Data file.
Similar articles
-
Genesis, Evolution and Prevalence of H5N6 Avian Influenza Viruses in China.
Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, Zhao L, Li M, Ding Z, Zou R, Xu W, Li H, Wang H, Tian K, Fu G, Huang Y, Shestopalov A, Li S, Xu B, Yu H, Luo T, Lu L, Xu X, Luo Y, Liu Y, Shi W, Liu D, Gao GF. Bi Y, et al. Cell Host Microbe. 2016 Dec 14;20(6):810-821. doi: 10.1016/j.chom.2016.10.022. Epub 2016 Dec 1. Cell Host Microbe. 2016. PMID: 27916476
-
Chen LJ, Lin XD, Guo WP, Tian JH, Wang W, Ying XH, Wang MR, Yu B, Yang ZQ, Shi M, Holmes EC, Zhang YZ. Chen LJ, et al. J Gen Virol. 2016 Apr;97(4):844-854. doi: 10.1099/jgv.0.000399. Epub 2016 Jan 12. J Gen Virol. 2016. PMID: 26758561
-
Yu X, Jin T, Cui Y, Pu X, Li J, Xu J, Liu G, Jia H, Liu D, Song S, Yu Y, Xie L, Huang R, Ding H, Kou Y, Zhou Y, Wang Y, Xu X, Yin Y, Wang J, Guo C, Yang X, Hu L, Wu X, Wang H, Liu J, Zhao G, Zhou J, Pan J, Gao GF, Yang R, Wang J. Yu X, et al. J Virol. 2014 Mar;88(6):3423-31. doi: 10.1128/JVI.02059-13. Epub 2014 Jan 8. J Virol. 2014. PMID: 24403589 Free PMC article.
-
Novel human H7N9 influenza virus in China.
Wang C, Luo J, Wang J, Su W, Gao S, Zhang M, Xie L, Ding H, Liu S, Liu X, Chen Y, Jia Y, He H. Wang C, et al. Integr Zool. 2014 Jun;9(3):372-5. doi: 10.1111/1749-4877.12047. Integr Zool. 2014. PMID: 24952971 Review.
-
Emergence and development of H7N9 influenza viruses in China.
Zhu H, Lam TT, Smith DK, Guan Y. Zhu H, et al. Curr Opin Virol. 2016 Feb;16:106-113. doi: 10.1016/j.coviro.2016.01.020. Epub 2016 Feb 26. Curr Opin Virol. 2016. PMID: 26922715 Review.
Cited by 23 articles
-
The molecular determinants of antigenic drift in a novel avian influenza A (H9N2) variant virus.
Zheng Y, Guo Y, Li Y, Liang B, Sun X, Li S, Xia H, Ping J. Zheng Y, et al. Virol J. 2022 Feb 5;19(1):26. doi: 10.1186/s12985-022-01755-9. Virol J. 2022. PMID: 35123509 Free PMC article.
-
Bhat S, James J, Sadeyen JR, Mahmood S, Everest HJ, Chang P, Walsh SK, Byrne AMP, Mollett B, Lean F, Sealy JE, Shelton H, Slomka MJ, Brookes SM, Iqbal M. Bhat S, et al. J Virol. 2022 Mar 9;96(5):e0185621. doi: 10.1128/jvi.01856-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019727 Free PMC article.
-
Li X, Zhang R, Huang Z, Yao D, Luo L, Chen J, Ye W, Li L, Xiao S, Liu X, Ou X, Sun B, Xu M, Yang R, Zhang X. Li X, et al. Food Environ Virol. 2022 Mar;14(1):30-39. doi: 10.1007/s12560-021-09506-9. Epub 2022 Jan 7. Food Environ Virol. 2022. PMID: 34997459
-
Reassortment Network of Influenza A Virus.
Gong X, Hu M, Chen W, Yang H, Wang B, Yue J, Jin Y, Liang L, Ren H. Gong X, et al. Front Microbiol. 2021 Dec 16;12:793500. doi: 10.3389/fmicb.2021.793500. eCollection 2021. Front Microbiol. 2021. PMID: 34975817 Free PMC article.
-
Yang H, Hu M, Wang B, Jin Y, Gong X, Liang L, Yue J, Chen W, Ren H. Yang H, et al. Front Microbiol. 2021 Dec 16;12:751142. doi: 10.3389/fmicb.2021.751142. eCollection 2021. Front Microbiol. 2021. PMID: 34975784 Free PMC article.
References
-
- OIE. Update on avian influenza in animals (types H5 and H7). https://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza (2019).
-
- Guo Y, Li J, Cheng X. Discovery of men infected by avian influenza A (H9N2) virus. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 1999;13:105–108. - PubMed
-
- WHO. Monthly risk assessment summary. https://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment... (2019).
Publication types
MeSH terms
LinkOut - more resources
-
Full Text Sources
-
Medical
