Flagellar Basal Body Structural Proteins FlhB, FliM, and FliY Are Required for Flagellar-Associated Protein Expression in Listeria monocytogenes - PubMed
doi: 10.3389/fmicb.2018.00208. eCollection 2018.
Hang Wang 1 , Tiantian Ma 1 , Xiao Han 1 , Yongchun Yang 1 , Jing Sun 1 , Zhongwei Chen 1 , Huifei Yu 1 , Yi Hang 1 , Fengdan Liu 1 , Weihuan Fang 1 2 , Lingli Jiang 3 , Chang Cai 1 4 , Houhui Song 1
Affiliations
- PMID: 29487588
- PMCID: PMC5816908
- DOI: 10.3389/fmicb.2018.00208
Free PMC article
Flagellar Basal Body Structural Proteins FlhB, FliM, and FliY Are Required for Flagellar-Associated Protein Expression in Listeria monocytogenes
Changyong Cheng et al. Front Microbiol. .
Free PMC article
Abstract
Listeria monocytogenes is a food-associated bacterium that is responsible for food-related illnesses worldwide. In the L. monocytogenes EGD-e genome, FlhB, FliM, and FliY (encoded by lmo0679, lmo0699, and lmo0700, respectively) are annotated as putative flagella biosynthesis factors, but their functions remain unknown. To explore whether FlhB, FliM, and FliY are involved in Listeria flagella synthesis, we constructed flhB, fliM, fliY, and other flagellar-related gene deletion mutants using a homologous recombination strategy. Then, we analyzed the motility, flagella synthesis, and protein expression of these mutant strains. Motility and flagella synthesis were completely abolished in the absence of flhB, fliM, or fliY. These impaired phenotypes were fully restored in the complemented strains CΔflhB, CΔfliM, and CΔfliY. The transcriptional levels of flagellar-related genes, including flaA, fliM, fliY, lmo0695, lmo0698, fliI, and fliS, were downregulated markedly in the absence of flhB, fliM, or fliY. Deletion of flhB resulted in the complete abolishment of FlaA expression, while it decreased FliM and FliY expression. The expression of FlaA was abolished completely in the absence of fliM or fliY. No significant changes were found in the expression of FlhF and two flagella synthesis regulatory factors, MogR and GmaR. We demonstrate for the first time that FlhB, FliM, and FliY not only mediate Listeria motility, but also are involved in regulating flagella synthesis. This study provides novel insights that increase our understanding of the roles played by FlhB, FliM, and FliY in the flagellar type III secretion system in L. monocytogenes.
Keywords: Listeria monocytogenes; flagella; motility; protein expression; type III secretion system.
Figures
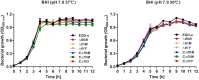
Growth of L. monocytogenes mutants in BHI medium at 37°C (A) and 30°C (B). Overnight cultures were resuspended in fresh BHI medium and then incubated at 37°C (A) and 30°C (B) for 12 h. Then, the OD600 nm was measured at 1-h intervals. The data are expressed as the mean ± standard deviation (SD) of three independent experiments.

FlhB, FliM, and FliY contribute to L. monocytogenes motility and flagellar formation. A motility assay (A,B) and TEM (C) were performed by growing the L. monocytogenes wild-type strain EGD-e, the mutants ΔflhB, ΔfliM and ΔfliY, and the complemented strains CΔflhB, CΔfliM, and CΔfliY on soft agar (0.25%) at 30°C or 37°C for 16 h. Scale: 1 μm.
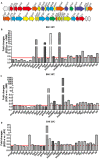
The flagellar-motility gene cluster in L. monocytogenes EGD-e (A) and the changes of the transcriptional levels of flagellar-related genes in the absence of flhB (B), fliM (C), or fliY (D). Relative quantification of flagellar-related gene mRNA levels in the L. monocytogenes wild-type strain EGD-e, the mutants ΔflhB, ΔfliM, and ΔfliY, and the complemented strains CΔflhB, CΔfliM, and CΔfliY at 30°C. Values are expressed as the mean ± SD. Solid lines indicate a 1.5-fold change in transcription of the genes of interest.
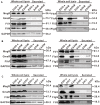
Changes in the levels of flagellar-related proteins in the absence of flhB (A), fliM (B), or fliY (C). Bacterial overnight cultures of the L. monocytogenes wild-type strain EGD-e, the ΔflhB, ΔfliM, ΔfliY mutants, and the complemented strains CΔflhB, CΔfliM, and CΔfliY were diluted 1:100 into 100 ml of fresh BHI broth and statically grown to stationary phase. Bacterial sediments and culture supernatants were collected to obtain the different cell fractions. Proteins were separated by 12% SDS-PAGE and immunoblotted with α-FlgG, α-MogR, α-FlhF, α-FliM, α-FliY, α-FlaA, α-FliS, or α-GmaR antisera. GAPDH was used as an internal control. The predicted molecular weight of each protein is indicated on the right.

Model for the roles of FlhB, FliY, and FliM in flagellar formation and bacterial motility. FlhB as an export switch factor is responsible for transporting intracellular proteins to the extracellular, including FlgG and FlaA. Besides its role in the flagellar switch complex, the C-ring is also involved in the export process, FliM and FliY are components of the C ring, their defects may lead to a variety of specific substrate proteins that can not be transported. FliS chaperone binds to filament (FlaA) in the cytoplasm and efficiently transfers FlaA to a sorting platform of the flagellar export apparatus during the flagellar filament assembly. The flhB, fliM, and fliY mutant strains are incapable of synthesizing filament, thus causing the bacteria to lose power and non-motile. FlhF is a GTPase associated with flagellin localization and flagellar transcription is still subject to temperature-dependent regulation of MogR and GmaR strictly, all these three regulators are not disturbed by FlhB, FliM, and FliY.
Similar articles
-
Structure and activity of the flagellar rotor protein FliY: a member of the CheC phosphatase family.
Sircar R, Greenswag AR, Bilwes AM, Gonzalez-Bonet G, Crane BR. Sircar R, et al. J Biol Chem. 2013 May 10;288(19):13493-502. doi: 10.1074/jbc.M112.445171. Epub 2013 Mar 26. J Biol Chem. 2013. PMID: 23532838 Free PMC article.
-
Gründling A, Burrack LS, Bouwer HG, Higgins DE. Gründling A, et al. Proc Natl Acad Sci U S A. 2004 Aug 17;101(33):12318-23. doi: 10.1073/pnas.0404924101. Epub 2004 Aug 9. Proc Natl Acad Sci U S A. 2004. PMID: 15302931 Free PMC article.
-
Kamp HD, Higgins DE. Kamp HD, et al. PLoS Pathog. 2011 Aug;7(8):e1002153. doi: 10.1371/journal.ppat.1002153. Epub 2011 Aug 4. PLoS Pathog. 2011. PMID: 21829361 Free PMC article.
-
Henderson LD, Matthews-Palmer TRS, Gulbronson CJ, Ribardo DA, Beeby M, Hendrixson DR. Henderson LD, et al. mBio. 2020 Jan 7;11(1):e02286-19. doi: 10.1128/mBio.02286-19. mBio. 2020. PMID: 31911488 Free PMC article.
-
How bacteria maintain location and number of flagella?
Schuhmacher JS, Thormann KM, Bange G. Schuhmacher JS, et al. FEMS Microbiol Rev. 2015 Nov;39(6):812-22. doi: 10.1093/femsre/fuv034. Epub 2015 Jul 20. FEMS Microbiol Rev. 2015. PMID: 26195616 Review.
Cited by 6 articles
-
Anuntakarun S, Sawaswong V, Jitvaropas R, Praianantathavorn K, Poomipak W, Suputtamongkol Y, Chirathaworn C, Payungporn S. Anuntakarun S, et al. Genomics Inform. 2021 Sep;19(3):e31. doi: 10.5808/gi.21037. Epub 2021 Sep 30. Genomics Inform. 2021. PMID: 34638178 Free PMC article.
-
Thai SN, Lum MR, Naegle J, Onofre M, Abdulla H, Garcia A, Fiterz A, Arnell A, Lwin TT, Kavanaugh A, Hikmat Z, Garabedian N, Ngo RT, Dimaya B, Escamilla A, Barseghyan L, Shibatsuji M, Soltani S, Butcher L, Hikmat F, Amirian D, Bazikyan A, Brandt N, Sarkisian M, Munoz X, Ovakimyan A, Burnett E, Pham JN, Shirvanian A, Hernandez R, Vardapetyan M, Wada M, Ramirez C, Zakarian M, Billi F. Thai SN, et al. J Bacteriol. 2021 Nov 5;203(23):e0029321. doi: 10.1128/JB.00293-21. Epub 2021 Sep 20. J Bacteriol. 2021. PMID: 34543106 Free PMC article.
-
Jia T, Liu B, Mu H, Qian C, Wang L, Li L, Lu G, Zhu W, Guo X, Yang B, Huang D, Feng L, Liu B. Jia T, et al. mBio. 2021 Mar 9;12(2):e03605-20. doi: 10.1128/mBio.03605-20. mBio. 2021. PMID: 33688013 Free PMC article.
-
Melian C, Castellano P, Segli F, Mendoza LM, Vignolo GM. Melian C, et al. Front Microbiol. 2021 Jan 29;12:604126. doi: 10.3389/fmicb.2021.604126. eCollection 2021. Front Microbiol. 2021. PMID: 33584610 Free PMC article.
-
Kragh ML, Truelstrup Hansen L. Kragh ML, et al. Front Microbiol. 2020 Jan 22;10:3132. doi: 10.3389/fmicb.2019.03132. eCollection 2019. Front Microbiol. 2020. PMID: 32038566 Free PMC article.
References
LinkOut - more resources
-
Full Text Sources
-
Other Literature Sources
-
Molecular Biology Databases
