Listeria monocytogenes 10403S Arginine Repressor ArgR Finely Tunes Arginine Metabolism Regulation under Acidic Conditions - PubMed
doi: 10.3389/fmicb.2017.00145. eCollection 2017.
Zhimei Dong 1 , Xiao Han 1 , Jing Sun 1 , Hang Wang 1 , Li Jiang 1 , Yongchun Yang 1 , Tiantian Ma 1 , Zhongwei Chen 1 , Jing Yu 1 , Weihuan Fang 2 , Houhui Song 1
Affiliations
- PMID: 28217122
- PMCID: PMC5291005
- DOI: 10.3389/fmicb.2017.00145
Free PMC article
Listeria monocytogenes 10403S Arginine Repressor ArgR Finely Tunes Arginine Metabolism Regulation under Acidic Conditions
Changyong Cheng et al. Front Microbiol. .
Free PMC article
Abstract
Listeria monocytogenes is able to colonize human and animal intestinal tracts and to subsequently cross the intestinal barrier, causing systemic infection. For successful establishment of infection, L. monocytogenes must survive the low pH environment of the stomach. L. monocytogenes encodes a functional ArgR, a transcriptional regulator belonging to the ArgR/AhrC arginine repressor family. We aimed at clarifying the specific functions of ArgR in arginine metabolism regulation, and more importantly, in acid tolerance of L. monocytogenes. We showed that ArgR in the presence of 10 mM arginine represses transcription and expression of the argGH and argCJBDF operons, indicating that L. monocytogenes ArgR plays the classical role of ArgR/AhrC family proteins in feedback inhibition of the arginine biosynthetic pathway. Notably, transcription and expression of arcA (encoding arginine deiminase) and sigB (encoding an alternative sigma factor B) were also markedly repressed by ArgR when bacteria were exposed to pH 5.5 in the absence of arginine. However, addition of arginine enabled ArgR to derepress the transcription and expression of these two genes. Electrophoretic mobility shift assays showed that ArgR binds to the putative ARG boxes in the promoter regions of argC, argG, arcA, and sigB. Reporter gene analysis with gfp under control of the argG promoter demonstrated that ArgR was able to activate the argG promoter. Unexpectedly, deletion of argR significantly increased bacterial survival in BHI medium adjusted to pH 3.5 with lactic acid. We conclude that this phenomenon is due to activation of arcA and sigB. Collectively, our results show that L. monocytogenes ArgR finely tunes arginine metabolism through negative transcriptional regulation of the arginine biosynthetic operons and of the catabolic arcA gene in an arginine-independent manner during lactic acid-induced acid stress. ArgR also appears to activate catabolism as well as sigB transcription by anti-repression in an arginine-dependent way.
Keywords: ArgR; Listeria monocytogenes; acid tolerance; arginine repressor; regulation.
Figures

Listeria monocytogenes ArgR protein is a member of ArgR/AhrC family transcriptional regulators. (A) Amino acid sequence alignment of L. monocytogenes ArgR against the members of the ArgR/AhrC family from Bacillus subtilis, B. stearothermophilus, Corynebacterium glutamicum, Escherichia coli, Mycobacterium tuberculosis, Streptomyces clavuligerus, and S. coelicolor. The two conserved motifs that are responsible for DNA and arginine binding are blackened. (B) Predicted tertiary fold of L. monocytogenes ArgR using the B. subtilis ArgR (PDB: 1F9N) as the template in the SWISS-MODEL Workspace. (C) SDS-PAGE analysis of glutaraldehyde crosslinking of L. monocytogenes ArgR for the identification of protein polymers. The monomeric, trimeric and hexameric proteins are indicated by arrows. (D) Promoter/operator elements containing binding sites of ArgR (ARG box) were identified by searching the L. monocytogenes genome with a position weight matrix derived from known ArgR recognition elements using the Virtual Footprint software program (as described in detail in the Materials and Methods). The identified potential consensus binding sites of ArgR in the promoter region from L. monocytogenes gene argC, argG, arcA, and sigB were further aligned with those from B. subtilis, B. licheniformis, and S. coelicolor.
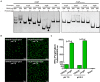
(A) ArgR binds in vitro to the operating sites of argC, argG, arcA and sigB, and activates the argG operon promoter. Gel mobility shift assay (EMSA) experiments were employed to examine binding of the recombinant ArgR from L. monocytogenes (ArgR) and its amino acid mutant protein (ArgRS42AR43A) to the argC, argG, arcA, and sigB promoter region DNA. The promoter fragments were obtained by PCR with primers specified in Supplementary Table S1, and incubated with recombinant proteins for 30 min at room temperature. Gel retardation by DNA–protein complexes was monitored after ethidium bromide staining. The housekeeping gene gyrB was used as a negative control (NC) for the EMSAs. Arrows indicate DNA-protein complexes. (B,C) ArgR activates the argG operon promoter. The fluorescence intensity was observed by confocal laser scanning microscopy of overnight grown L. monocytogenes wild-type and ArgR mutant strains carrying a gfp3 reporter fused with the promoter of argG, and then stress-treated for an additional 1 h under pH 7.0 and 5.5 conditions (B). Bars represent the relative fluorescence units (RFU) after subtracting the absolute values for the PBS control (C). Data shown represents the Mean ± SD of three independent experiments, each performed in duplicate. ∗∗P < 0.01 for comparisons between the wild-type and mutant strains.

ArgR regulates the transcription of argC, argG, arcA, and sigB using arginine as a cofactor. Relative quantification of argR, arcA, sigB, argC, and argG mRNA and protein expression levels in L. monocytogenes wild-type and ΔArgR mutant strains under different pH conditions (7.0 and 5.5) in the presence (A–C) or absence (D–F) of exogenous arginine (10 mM). Values are expressed as Mean ± SD. The dotted lines indicate the onefold change in transcription of the interest genes.
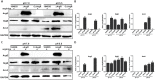
ArgR regulates the expression of ArgG, SigB, and ArcA using arginine as a cofactor. Total bacterial cell-free protein was isolated 2 h after stress under different pH conditions (7.0 and 5.5) in the absence (A) or presence (C) of exogenous arginine (10 mM), and the protein expression levels of ArgG, SigB, and ArcA were then determined by Western blotting. GAPDH was used as an internal control. The results are indicated as of the gray scale ratio of each interest protein to GADPH (B,D). Data shown represents the Mean ± SD of two independent experiments. ∗∗P < 0.01; ns means no significance.

Deletion of ArgR enhances survival of L. monocytogenes in the lethal acidic conditions. Overnight-grown L. monocytogenes wild-type and mutant strains were harvested, washed and then incubated in BHI broth (pre-adjusted to pH 3.5 using 3 M lactic acid, LA) at 37°C. Survivors were enumerated at regular intervals by plating serial dilutions on BHI plate. Data are expressed as Mean ± SD of recovery rate for each strain. ∗P < 0.05; ∗∗P < 0.01; ns means no significance.
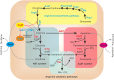
Schematic representation of regulatory mechanism employed by ArgR in L. monocytogenes. L. monocytogenes ArgR plays a classical role of ArgR/AhrC family in feedback inhibitory of arginine biosynthetic pathway (highlighted in yellow) using arginine as a cofactor. ArgR unexpectedly represses the transcription and expression of arcA and sigB in the absence of exogenous arginine, preventing arginine degradation via the ADI and AgDI pathways (highlighted in light cyan and light red, respectively). Addition of arginine leads to derepression of arcA and sigB, allowing utilization of arginine as a nitrogen and energy source via the arginine degradation pathway.
Similar articles
-
Interaction between ArgR and AhrC controls regulation of arginine metabolism in Lactococcus lactis.
Larsen R, Kok J, Kuipers OP. Larsen R, et al. J Biol Chem. 2005 May 13;280(19):19319-30. doi: 10.1074/jbc.M413983200. Epub 2005 Mar 4. J Biol Chem. 2005. PMID: 15749710
-
Yim SH, Jung S, Lee SK, Cheon CI, Song E, Lee SS, Shin J, Lee MS. Yim SH, et al. J Ind Microbiol Biotechnol. 2011 Dec;38(12):1911-20. doi: 10.1007/s10295-011-0977-9. Epub 2011 May 11. J Ind Microbiol Biotechnol. 2011. PMID: 21559975
-
Listeria monocytogenes ArcA contributes to acid tolerance.
Cheng C, Chen J, Shan Y, Fang C, Liu Y, Xia Y, Song H, Fang W. Cheng C, et al. J Med Microbiol. 2013 Jun;62(Pt 6):813-821. doi: 10.1099/jmm.0.055145-0. Epub 2013 Mar 21. J Med Microbiol. 2013. PMID: 23518652
-
Expression and characterization of ArgR, an arginine regulatory protein in Corynebacterium crenatum.
Chen XL, Zhang B, Tang L, Jiao HT, Xu HY, Xu F, Xu H, Wei H, Xiong YH. Chen XL, et al. Biomed Environ Sci. 2014 Jun;27(6):436-43. doi: 10.3967/bes2014.072. Biomed Environ Sci. 2014. PMID: 24961853
-
ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis.
Larsen R, Buist G, Kuipers OP, Kok J. Larsen R, et al. J Bacteriol. 2004 Feb;186(4):1147-57. doi: 10.1128/JB.186.4.1147-1157.2004. J Bacteriol. 2004. PMID: 14762010 Free PMC article.
Cited by 14 articles
-
The arginine deaminase system plays distinct roles in Borrelia burgdorferi and Borrelia hermsii.
Richards CL, Raffel SJ, Bontemps-Gallo S, Dulebohn DP, Herbert TC, Gherardini FC. Richards CL, et al. PLoS Pathog. 2022 Mar 14;18(3):e1010370. doi: 10.1371/journal.ppat.1010370. eCollection 2022 Mar. PLoS Pathog. 2022. PMID: 35286343 Free PMC article.
-
Landscape of Stress Response and Virulence Genes Among Listeria monocytogenes Strains.
Lakicevic BZ, Den Besten HMW, De Biase D. Lakicevic BZ, et al. Front Microbiol. 2022 Jan 20;12:738470. doi: 10.3389/fmicb.2021.738470. eCollection 2021. Front Microbiol. 2022. PMID: 35126322 Free PMC article. Review.
-
Quereda JJ, Morón-García A, Palacios-Gorba C, Dessaux C, García-Del Portillo F, Pucciarelli MG, Ortega AD. Quereda JJ, et al. Virulence. 2021 Dec;12(1):2509-2545. doi: 10.1080/21505594.2021.1975526. Virulence. 2021. PMID: 34612177 Free PMC article.
-
Papiran R, Hamedi J. Papiran R, et al. Appl Biochem Biotechnol. 2021 Nov;193(11):3425-3441. doi: 10.1007/s12010-021-03609-6. Epub 2021 Jul 1. Appl Biochem Biotechnol. 2021. PMID: 34196920
-
Chen YY, Kommadath A, Vahmani P, Visvalingam J, Dugan MER, Yang X. Chen YY, et al. Appl Environ Microbiol. 2021 Apr 13;87(9):e03027-20. doi: 10.1128/AEM.03027-20. Print 2021 Apr 13. Appl Environ Microbiol. 2021. PMID: 33608290 Free PMC article.
References
LinkOut - more resources
-
Full Text Sources
-
Other Literature Sources
