Deletion of glutaredoxin promotes oxidative tolerance and intracellular infection in Listeria monocytogenes - PubMed
. 2019 Dec;10(1):910-924.
doi: 10.1080/21505594.2019.1685640.
Yi Hang 1 , Yue Han 1 , Xian Zhang 1 , Li Gan 1 , Chang Cai 1 2 , Zhongwei Chen 1 , Yang Yang 1 , Quanjiang Song 1 , Chunyan Shao 1 , Yongchun Yang 1 , Yingshan Zhou 1 , Xiaodu Wang 1 , Changyong Cheng 1 , Houhui Song 1
Affiliations
- PMID: 31680614
- PMCID: PMC6844310
- DOI: 10.1080/21505594.2019.1685640
Free PMC article
Deletion of glutaredoxin promotes oxidative tolerance and intracellular infection in Listeria monocytogenes
Jing Sun et al. Virulence. 2019 Dec.
Free PMC article
Abstract
Thiol-disulfide glutaredoxin systems of bacterial cytoplasm favor reducing conditions for the correct disulfide bonding of functional proteins, and therefore were employed by bacteria to defend against oxidative stress. Listeria monocytogenes has been shown to encode a putative glutaredoxin, Grx (encoded by lmo2344), while the underlying roles remain unknown. Here we suggest an unexpected role of L. monocytogenes Grx in oxidative tolerance and intracellular infection. The recombinant Grx was able to efficiently catalyze the thiol-disulfide oxidoreduction of insulin in the presence of DTT as an election donor. Unexpectedly, the deletion of grx resulted in a remarkably increased tolerance and survival ability of this bacteria when exposed to various oxidizing agents, including diamide, and copper and cadmium ions. Furthermore, loss of grx significantly promoted bacterial invasion and proliferation in human epithelial Caco-2 cells and murine macrophages, as well as a notably increasing invasion but not cell-to-cell spread in the murine fibroblasts L929 cells. More importantly, L. monocytogenes lacking the glutaredoxin exhibited more efficient proliferation and recovery in the spleens and livers of the infected mice, and hence became more virulent by upregulating the virulence factors, InlA and InlB. In summary, we here for the first time demonstrated that L. monocytogenes glutaredoxin plays a counterintuitive role in bacterial oxidative resistance and intracellular infection, which is the first report to provide valuable evidence for the role of glutaredoxins in bacterial infection, and more importantly suggests a favorable model to illustrate the functional diversity of bacterial Grx systems during environmental adaption and host infection.
Keywords: Listeria monocytogenes; glutaredoxin; intracellular infection; oxidative tolerance; oxidoreduction.
Figures
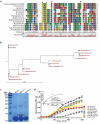
L. monocytogenes encodes a putative glutaredoxin, Grx that can catalyze reduction of insulin in the presence of DTT as an electron donor. (a) Amino acid sequence alignment of L. monocytogenes putative glutaredoxin (lmo2344) against homologues from L. innocua, L. grayi, B. subtilis, P. aeruginosa, S. flexneri, S.typhimurium, V.cholerae, K.pneumoniae,and P. multocida. The conserved CXXC catalytic motifs are denoted with asterisks. (b) Phylogenetic tree of L. monocytogenes putative glutaredoxin and homologs from the above bacterial species. The tree was constructed with the Neighbor-Joining (NJ) program and a bootstrap test of 100 replicates used to estimate the confidence of branching patterns, where the numbers on internal nodes represent the support values. (c) SDS-PAGE analysis of the purified histidine-tagged recombinant Grx and its site-directed mutants (GrxCPYC and GrxCGFS) overexpressed in E. coli. (d) In vitro insulin disulfide reductase activity of recombinant Grx and its site-directed mutants (GrxCPYC and GrxCGFS). The rate of insulin reduction was assessed by measuring the turbidity at 650 nm in reaction assays containing 1 mM DTT in the presence of 10–100 μM redoxins. The inset shows the Michaelis-Menten plot and kinetic parameters, Km, Vmax, and kcat, for Grx. L. monocytogenes thioredoxin A, TrxA that has efficient thiol-disulfide oxidoreduction activity, was taken as a reference control in this assay. Data are expressed as means ± SDs.
Deletion of grx did not affect bacterial in vitro growth and motility, but slightly decreased the bacterial colony size. (a-b) In vitro growth and colony size assay of L. monocytogenes wild-type EGD-e, and gene deletion and complementation mutants (Δgrx and CΔgrx). Overnight-grown bacteria were washed and diluted (1:100) in fresh BHI broth (a) or BHI agar medium (b), and incubated at 37°C for 12 hours. Kinetic growth at OD600 nm (a) was measured at 1-h intervals, and bacterial CFU numbers (b) counted at 4-h intervals. Data are expressed as means ± SDs. (c) The bacterial colony morphology grown on the BHI agar plates for 12 hours were observed by using a stereo-microscope. The size of the single bacterial colony was measured of 100 bacteria and data are expressed means ± SDs. **P < 0.01. (d) Bacterial flagellar formation observed by transmission electron microscopy (TEM) were performed on soft agar (0.25%) at 30°C.

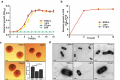
Deletion of grx remarkably increased tolerance to the oxidizing oxidants, copper, cadmium and diamide, but not to the hydrogen peroxide. (a-d) Growth of L. monocytogenes wild-type EGD-e, and gene deletion and complementation mutants (Δgrx and CΔgrx) in BHI agar medium supplemented with or without oxidative agents. Overnight-grown bacteria were subjected to hydrogen peroxide (a), copper (b), cadmium (c), and diamide (d) by spotting 108 CFU of each bacterial strain onto BHI plates containing various concentrations of oxidizingoxidants.
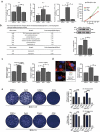
Mutation of grx increased the effciency of bacterial intracellular growth and cell-to-cell spreading via upregulating the internalins, InlA and InlB. (a) Intracellular growth of L. monocytogenes in murine-derived J774 macrophages. Gentamycin (50 μg/mL) was added 30 min post infection. The J774 cells infected with L. monocytogenes wild-type EGD-e and grx mutant were lysed at the indicated time points (2, 6, and 12 h), and viable bacteria serially plated on BHI plates. The number of recovered bacteria able to invade cells and survive are expressed as means ± SDs for each strain. (b) The transcriptional level changes of the virulence-associated factors (prfA, plcA, plcB, hly, mpl, inlA and inlB) of L. monocytogenes wild-type EGD-e and grx mutant, which were identified by the transcript analysis. Expression of InlB in the wild-type and mutant strains were assayed by Western blotting. (c) Adhesion and invasion of L. monocytogenes in human epithelial cells, Caco-2. Cells infected with L. monocytogenes wild-type EGD-e, and gene deletion and complementation mutants (Δgrx and CΔgrx) at the indicated time points were lysed and viable bacteria serially plated on BHI plates. The number of recovered bacteria able to invade cells and survive are expressed as means ± SDs for each strain. (d) Intracellular multiplication of L. monocytogenes in Caco-2 cells 6 h post infection. Bacteria were detected with anti-Lm (green), and bacteria actin tails and host actin were detected using phalloidin (red), while the cell nucleus was labeled with DAPI (blue). The scale bar is 10 μm. The high-magnification images displayed at the bottom of each image show F-actin (red), bacteria (green), and nuclei (blue). (e) Plaque assay performed on the L929 fibroblast monolayers infected by L. monocytogenes wild-type EGD-e, and gene deletion and complementation mutants (Δgrx and CΔgrx). The plaque numbers of the mutant strains were indicated as a percentage of those formed by the wild-type strain. The mutant strain hly, which is completely unable to spread during cell infection, was taken as a reference negative control. *P<0.05; **P<0.01; ns means no significance. Data are expressed as means ± SDs.
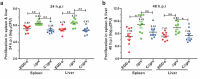
Deletion of grx resulted in the increased virulence in mice. Proliferation of L. monocytogenes in mice organs. The L. monocytogenes wild-type and mutant Δgrx bacteria were inoculated intraperitoneally into ICR mice at ~4 × 106 CFU. Animals were euthanized 24 (a) and 48 (b) hours post infection, and organs (livers and spleens) were recovered and homogenized. Homogenates were serially diluted and plated on BHI agar. Numbers of bacteria colonized in liver and spleen are expressed as means ± SDs of the log10CFU per organ for each group. *P<0.05; **P<0.01; ns means no significance.

A proposed model for L. monocytogenes Grx as a novel glutaredoxin in bacterial oxidative tolerance. (a) Based on sequence alignment using the full-length sequences of PerRs from B. subtilis and S. aureus as templates, four cysteine residues (C98, C101, C138 and C141) that forms a high-affinity Zn2+ binding site (Cys4Zn), and five other residues (H39, D87, H93, H95 and D106) that preferentially binds Fe2+ or Mn2+ are found to be perfectly conserved in L. monocytogenes PerR. These residues are indicated with asterisks. (b) Therefore, we here proposed that L. monocytogenes repressor PerR could be irreversibly inactivated when bacteria exposed to oxidative conditions and failed to switch back to the activated form in the absence of Grx, resulting in derepression of the PerR-regulated oxidizing response genes.
Similar articles
-
Cheng C, Dong Z, Han X, Wang H, Jiang L, Sun J, Yang Y, Ma T, Shao C, Wang X, Chen Z, Fang W, Freitag NE, Huang H, Song H. Cheng C, et al. Front Cell Infect Microbiol. 2017 Jun 28;7:287. doi: 10.3389/fcimb.2017.00287. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28702378 Free PMC article.
-
Cheng C, Wang X, Dong Z, Shao C, Yang Y, Fang W, Fang C, Wang H, Yang M, Jiang L, Zhou X, Song H. Cheng C, et al. Sci Rep. 2015 Nov 27;5:17370. doi: 10.1038/srep17370. Sci Rep. 2015. PMID: 26610705 Free PMC article.
-
Relative Roles of Listeriolysin O, InlA, and InlB in Listeria monocytogenes Uptake by Host Cells.
Phelps CC, Vadia S, Arnett E, Tan Y, Zhang X, Pathak-Sharma S, Gavrilin MA, Seveau S. Phelps CC, et al. Infect Immun. 2018 Sep 21;86(10):e00555-18. doi: 10.1128/IAI.00555-18. Print 2018 Oct. Infect Immun. 2018. PMID: 30061379 Free PMC article.
-
[Contribution of sigma B to environmental stress tolerance in Listeria monocytogenes--a review].
Zhang Q, Feng Y, Zhou Q, Luo Q, Zhang X, Qin L. Zhang Q, et al. Wei Sheng Wu Xue Bao. 2009 Oct;49(10):1282-8. Wei Sheng Wu Xue Bao. 2009. PMID: 20069872 Review. Chinese.
-
Virulence characterization of Listeria monocytogenes.
Reddy S, Lawrence ML. Reddy S, et al. Methods Mol Biol. 2014;1157:157-65. doi: 10.1007/978-1-4939-0703-8_13. Methods Mol Biol. 2014. PMID: 24792556 Review.
Cited by 5 articles
-
Kho ZY, Azad MAK, Han ML, Zhu Y, Huang C, Schittenhelm RB, Naderer T, Velkov T, Selkrig J, Zhou QT, Li J. Kho ZY, et al. PLoS Pathog. 2022 Mar 1;18(3):e1010308. doi: 10.1371/journal.ppat.1010308. eCollection 2022 Mar. PLoS Pathog. 2022. PMID: 35231068 Free PMC article.
-
Nguyen Huu C, Rai R, Yang X, Tikekar RV, Nitin N. Nguyen Huu C, et al. Ultrason Sonochem. 2021 Jun;74:105567. doi: 10.1016/j.ultsonch.2021.105567. Epub 2021 Apr 16. Ultrason Sonochem. 2021. PMID: 33957369 Free PMC article.
-
Li T, Wang Z, Han H, Teng D, Mao R, Hao Y, Yang N, Wang X, Wang J. Li T, et al. Appl Microbiol Biotechnol. 2021 Mar;105(6):2351-2361. doi: 10.1007/s00253-021-11176-3. Epub 2021 Feb 26. Appl Microbiol Biotechnol. 2021. PMID: 33635357
-
Cheng C, Han X, Xu J, Sun J, Li K, Han Y, Chen M, Song H. Cheng C, et al. Gut Microbes. 2021 Jan-Dec;13(1):1-19. doi: 10.1080/19490976.2021.1884517. Gut Microbes. 2021. PMID: 33573432 Free PMC article.
-
Zhang X, Guan C, Hang Y, Liu F, Sun J, Yu H, Gan L, Zeng H, Zhu Y, Chen Z, Song H, Cheng C. Zhang X, et al. Microorganisms. 2020 Jan 13;8(1):110. doi: 10.3390/microorganisms8010110. Microorganisms. 2020. PMID: 31941013 Free PMC article.
References
-
- Fahey RC. Glutathione analogs in prokaryotes. Biochim Biophys Acta. 2013;1830:3182–3198. - PubMed
-
- Van Laer K, Hamilton CJ, Messens J. Low-molecular-weight thiols in thiol-disulfide exchange. Antioxid Redox Signal. 2013;18:1642–1653. - PubMed
-
- Ezraty B, Gennaris A, Barras F, et al. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol. 2017;15:385–396. - PubMed
-
- Radoshevich L, Cossart P. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol. 2018;16:32–46. - PubMed
Publication types
MeSH terms
Substances
Grant support
This work was supported by National Natural Science Foundation of China (Nos. 31872620, 31770040, 31972648, 31802258 and 31502034), Zhejiang Provincial Natural Science Foundation (Nos. LY17C180001, LZ19C180001, LQ19C180002, LQ15C180002 and LQ19C180003), National Key Research and Development Program of China (2017YFC1200500), and Key Research & Development Program of Zhejiang Province (No. 2019C02052). The founders had no role in design of the study or analysis and interpretation of the data.
LinkOut - more resources
-
Full Text Sources
-
Other Literature Sources
-
Medical
